Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors
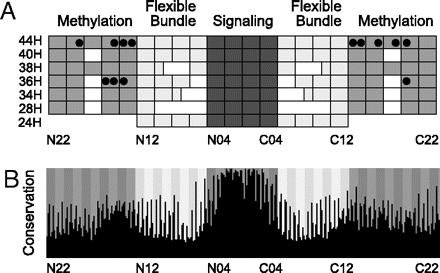 Schematic representation of MCP_CD features revealed by the multiple sequence alignment. The complete alignment is shown in SI Dataset 1. (A) Subdomain structure of major domain classes. The three subdomains, methylation helices, flexible bundle, and signaling, are indicated by medium, light, and dark gray, respectively. Each rectangle represents a group of two heptads. Heptads are numbered from N22 at the N terminus down to N01 at the center and then up from C01 at the center to C22 at the C terminus. This naming convention has the advantage that N- and C-terminal heptads with the same number are adjacent in the structure. Gap locations are shown in white. MCP classes are named 24H through 44H indicating the number of heptads (H). Experimentally determined methylation sites in class 36H MCPs from E. coli (28) and class 44H MCPs from B. subtilis (33, 40), H. salinarum (41, 42), and T. maritima (29, 43) are indicated by black circles. (B) Amino acid conservation within the MCP_CD. Position of each of 309 residues in the alignment is indicated by black columns. The column height shows the conservation level (see SI Text for details). Positions of the 44 seven-residue heptads (N22–C22) are indicated by background gray shading. The first and fourth (a and d) residues of each heptad are strongly conserved.
Schematic representation of MCP_CD features revealed by the multiple sequence alignment. The complete alignment is shown in SI Dataset 1. (A) Subdomain structure of major domain classes. The three subdomains, methylation helices, flexible bundle, and signaling, are indicated by medium, light, and dark gray, respectively. Each rectangle represents a group of two heptads. Heptads are numbered from N22 at the N terminus down to N01 at the center and then up from C01 at the center to C22 at the C terminus. This naming convention has the advantage that N- and C-terminal heptads with the same number are adjacent in the structure. Gap locations are shown in white. MCP classes are named 24H through 44H indicating the number of heptads (H). Experimentally determined methylation sites in class 36H MCPs from E. coli (28) and class 44H MCPs from B. subtilis (33, 40), H. salinarum (41, 42), and T. maritima (29, 43) are indicated by black circles. (B) Amino acid conservation within the MCP_CD. Position of each of 309 residues in the alignment is indicated by black columns. The column height shows the conservation level (see SI Text for details). Positions of the 44 seven-residue heptads (N22–C22) are indicated by background gray shading. The first and fourth (a and d) residues of each heptad are strongly conserved.Abstract
As an important model for transmembrane signaling, methyl-accepting chemotaxis proteins (MCPs) have been extensively studied by using genetic, biochemical, and structural techniques. However, details of the molecular mechanism of signaling are still not well understood. The availability of genomic information for hundreds of species enables the identification of features in protein sequences that are conserved over long evolutionary distances and thus are critically important for function. We carried out a large-scale comparative genomic analysis of the MCP signaling and adaptation domain family and identified features that appear to be critical for receptor structure and function. Based on domain length and sequence conservation, we identified seven major MCP classes and three distinct structural regions within the cytoplasmic domain: signaling, methylation, and flexible bundle subdomains. The flexible bundle subdomain, not previously recognized in MCPs, is a conserved element that appears to be important for signal transduction. Remarkably, the N- and C-terminal helical arms of the cytoplasmic domain maintain symmetry in length and register despite dramatic variation, from 24 to 64 7-aa heptads in overall domain length. Loss of symmetry is observed in some MCPs, where it is concomitant with specific changes in the sensory module. Each major MCP class has a distinct pattern of predicted methylation sites that is well supported by experimental data. Our findings indicate that signaling and adaptation functions within the MCP cytoplasmic domain are tightly coupled, and that their coevolution has contributed to the significant diversity in chemotaxis mechanisms among different organisms.